Affiliation searches: the Why, What, and How of our Canonical Affiliation Feature
Carolyn Stern Grant and Matthew Templeton (ADS)
15 Jan 2020
One of the features new to ADS since the new interface has launched is search and organization by institutional affiliation. The 15 million publications in ADS have more than 35 million combined author affiliations. The ADS has long wanted to have these data in a searchable format, and we introduced a new curated affiliation feature in early 2019. The project involved matching existing publisher-provided affiliation strings to unique, curated affiliation identifiers and institution strings, stored in an internal affiliation database, and constructing a pipeline to match the publisher-provided affiliation strings in incoming new publications to the appropriate entry in this database. Constructing the internal affiliation database took a lot of manual labor – in fact, much more than the pipeline that resolves the affiliation strings – and while this database will never be 100 percent complete, we’re hoping to use machine learning techniques to reduce the amount of human effort required to match affiliation data to their identifiers.
Users are reminded that while affiliation information is largely complete for recent refereed literature, not all records contain an affiliation, therefore, searching by affiliation alone will inherently be incomplete. We strongly recommend combining affiliation searches with author searches for best results.
Using affiliations as part of your search strategy: Author searches and the Affiliation facet
Affiliation data for papers are available via the “Affiliations” facet in ADS. As an example, if you search for a first author, you’ll get a list of papers having that first author, and on the left-hand side of the results page you’ll see ways to refine those results – who their coauthors are, whether the paper is in the astronomy, physics, or general database, whether it’s a refereed publication or not, and so on. One of those facets is “Affiliations”, and by clicking it you’ll get a list of institutional affiliations sorted by the number of papers having that affiliation.
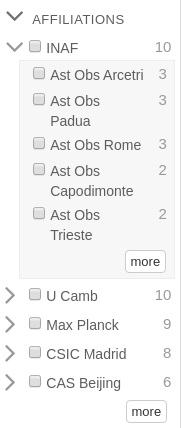
Example of the Affiliations facet
Using one of this blog’s coauthors, searching for first_author:“Templeton, Matthew” brings up a list of his papers along with a few other “Matthew Templeton”s who are active researchers in ADS-relevant fields. Selecting the “astronomy” collection and “limit to” yields (mostly) just his set of first-authored publications, and refereed lowers it further to a manageable number. If you then open the Affiliations facet, you’re presented with a list of all affiliations (of any author) contained within his papers. Matthew Templeton’s historical affiliations – AAVSO, New Mexico State University, Yale University, Los Alamos National Laboratory – are among the most common in the list. However, you also find affiliations of co-authors too, for example Iowa State and the Center for Astrophysics.
Using affiliations as part of your search strategy: Direct affiliation searches
Affiliations in the ADS have been indexed in several different fields, with the intention of allowing multiple use cases. We have currently assigned identifiers with parent/child relationships, such as an academic department within a university. A child may have multiple parents, but we restrict a child from having children of its own. This has required a few modifications to remain useful. For instance, so that University of California schools can identify departments, we have assigned them a parent status, even though the “University of California System” should really be the parent level. Likewise, NASA’s Goddard Space Flight Center is at a parent level, as are France’s CNRS institutions to allow for further subdivision. Further work on a schema to allow more complex relationships between institutions is under development in conjunction with work by the ROR Community.
We’ve recently changed the way we index affiliations by introducing a new search field: “affil”. Affil combines all of the available affiliation data – raw strings, canonical strings, IDs, and abbreviations into a single, searchable field. It’s intended to be a more comprehensive search of both raw strings and the enriched institution asssignments we’ve applied to our data. So for example, searching for affil:“UCB” will return papers where ‘UCB’ matches some part of the raw affiliation, but also return affiliations we’ve matched to the University of California at Berkeley. Because “UCB” is used by some authors to abbreviate other universities (for example “Catholic University of Brasilia, Brazil” or “University of Colorado, Boulder”), the affil search also finds these, but they can be de-selected in the affiliation facet.
In addition to this new field, we’ve maintained the original search terms for affiliations that we deployed last year:
-
aff: raw affiliation string, searchable word-by-word
-
aff_id: a string containing one or more of the affiliation IDs listed in our mapping of organizations to identifiers. This field will soon also accept 9-digit ROR IDs.
-
inst: the abbreviated institution name (e.g. “U Adelaide”) listed in our mapping of organizations to identifers.
So, you could search for aff:Harvard and get back all affiliations that contain Harvard in the affiliation string – including “Harvard Street”. To ensure you get only the University, you could search for aff:“Harvard University” but that would return only affiliation strings with that exact phrase, excluding “Harvard Univ.”, “Harvard U”, etc. Better would be to use the identifier for Harvard University by searching aff_id:A00211. This returns all affiliations which contain Harvard University at the parent level. Best yet would be to search by institution, inst:“Harvard U” which returns affiliations which contain Harvard University at the parent level, plus all affiliations for all of Harvard University’s children.
How we got here: the curation and pipeline process
The human element: curation
Production of the initial affiliation database began with two human-generated projects: creating a set of institutional identifiers, and matching affiliation metadata to these identifiers as precisely as possible. Both of these were done almost entirely by ADS Lead Curator Carolyn Grant.
There are hundreds of thousands of organizations around the world, and in principle any institution or business can be assigned an identifier. The overwhelming majority of these are not relevant to ADS, so the first task was to establish a list of institutions that match affiliations in our metadata. The current list stands at about 6600 departments, organizations, and parent institutions in all geographic regions of the world. Many of these institutions are also tagged with a second identifier, namely the identifier of the parent institution. For example, the parent institution of the Department of Physics at the University of California at Irvine is “University of California at Irvine”. For now, our affiliations have two levels only, so in this example, we do not assign “The University of California System” as the parent of UC Irvine, but our list of identifiers includes System identifiers in cases of ambiguity (e.g. a publisher-provided affiliation “Univ. of Cal.” with no city or other unambiguous identifying information).
The second, and by far most time-consuming, part of this process was the initial classification of millions of individual affiliation strings from the ADS bibliographic database. This involved both extraction of the author affiliation strings from the database and identification of the institution represented by that string.
As an example, a typical string might be “Physics Dept., UC Irvine” which would correspond to the affiliation of “Department of Physics, University of California at Irvine” and then assigning this string its corresponding alpha-numeric ID. The process sounds straightforward, but extraction of all strings relating to “Department”, “Physics”, “University of California”, and “Irvine” can be long and tedious, especially if you’re searching thousands of possible strings. There are ways to speed that process (e.g. by searching for “Irvine” and “Physics” in all strings, and then looking through just that set of results), but at the beginning it was an entirely human-curated process. Complicating things is the fact that many authors are affiliated with more than one institution, for example “Astronomy Dept., U. Texas – Austin, and UNAM-Morelia”. Publishers – especially smaller publications and conference proceedings – rarely use standardized text for affiliations, and even more rarely list multiple affiliations in separate entities in their metadata. So the curation process also involves splitting those multiple affiliations and then characterizing each affiliation separately. That process was repeated over and over for the 6600 affiliations that each occasionally had thousands or tens of thousands of different non-unique publisher-provided strings.
Cleaning the data turned out to be more of an art than a science. There exist curation tools for cleaning up messy data – the one we used almost exclusively was Open Refine (called Google Refine at the time). This allowed bulk substitutions, expansions, translations and more. But cleaning is not enough. For example, changing all instances of “UC Irvine” to “University of California Irvine” may seem like a good idea, except that new instances of “UC Irvine” are likely to continue coming in.
Automated Pipelines: assigning identifiers to specific author-affiliation-bibcode entities
With the initial set of affiliation strings identified we, along with ADS backend programmer Stephen McDonald, designed an automated pipeline to add canonical affiliation identifiers to our database of 15 million references. It’s a simple process. First we normalize all of the strings with all uppercase letters, remove spaces and a subset of punctuation marks not needed for disambiguation, which results in a reduced number of matching strings; often, strings will be identical without punctuation, such as strings with “… U.S.A.” and “… USA”. We then assign the strings to a dictionary, where the strings themselves are the keys, and the identifiers are the values. If a normalized incoming string matches a key in the dictionary, we assign it the appropriate aff_id, and the record with augmented affiliation data is sent back to our database of metadata for use in the next reindexing process.
We’ve entirely automated the process, so once the dictionary of affiliation strings and matched identifiers is created, it’s a hands-off component of our metadata processing pipeline. We update dictionaries about once a month, and occasionally more often if we have a large batch of incoming unmatched affiliation strings. This process is similar to the first curation step, with the only difference being that we prioritize strings according to how many times they appear. For example, if we receive a number of papers from collaborations with hundreds or thousands of authors and their affiliations don’t exactly match our dictionary, we need to add them. Early on in this process, we came across strings that had many thousands (and sometimes tens of thousands) of occurrences, but the latest batch of unclassified strings appear fewer than 550 times in our entire database. This maximum frequency drops each time we create a new dictionary.
However, we’re getting close to the limit of what we can do with human classification without lots of effort. Our most recent pass through our metadata has just over 4.8 million unique unclassified strings yielding a total of 16.6 million unclassified affiliations. There are about 6000 unclassified affiliations that appear 100 times or more in the metadata and assigning them aff_ids would provide a lot of new information. However, over 65 percent of unclassified affiliations occur fewer than 10 times, and 13 percent of unmatched affiliation strings only appear once in all of our affiliation metadata.
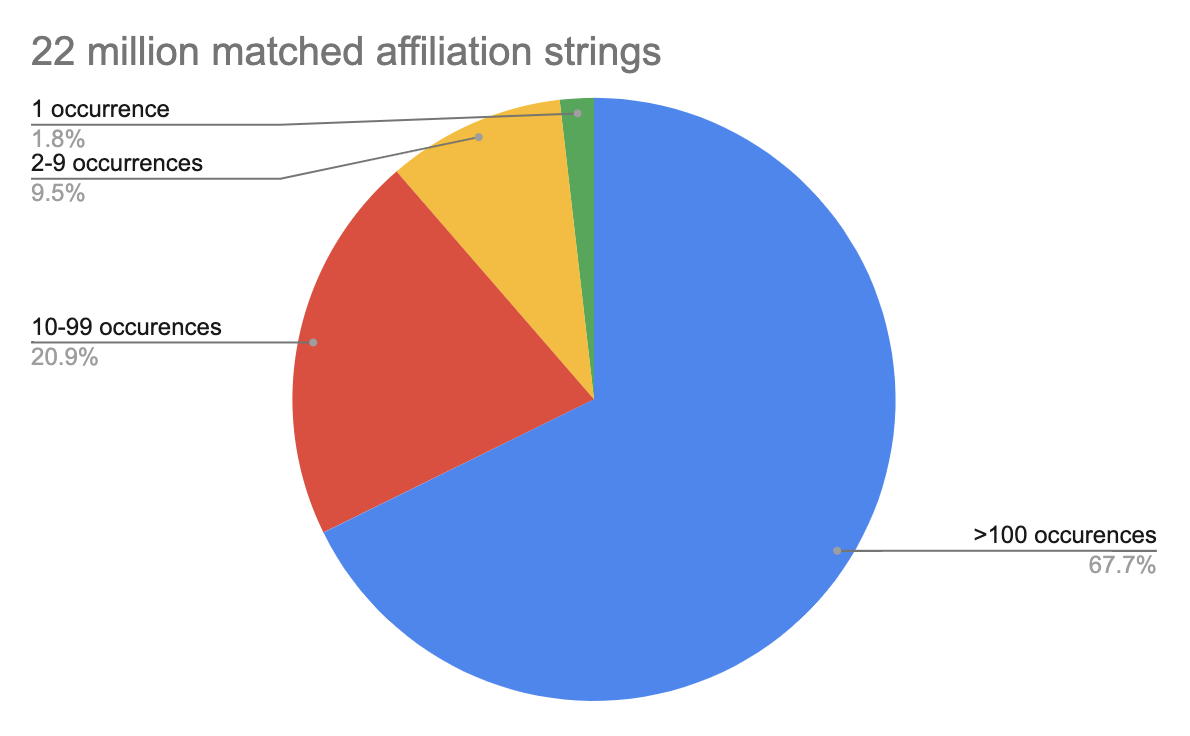
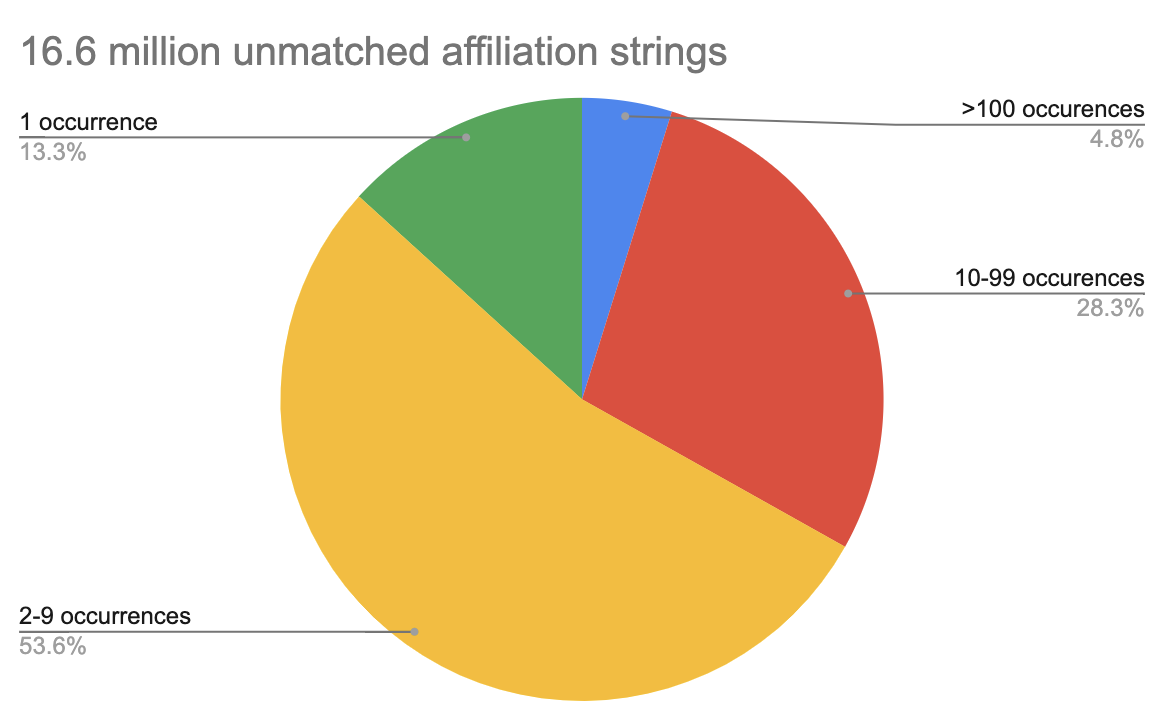 Left: Nearly 90 percent of matched publisher-provided affiliation strings in our database appear at least 10 times. Right: The number of unmatched affiliation strings is dominated by those appearing fewer than 10 times.
Left: Nearly 90 percent of matched publisher-provided affiliation strings in our database appear at least 10 times. Right: The number of unmatched affiliation strings is dominated by those appearing fewer than 10 times.
This is too much to work through by hand, but it represents valid affiliation data for millions of author-affiliation pairs. For the unmatched affiliations that occur fewer than 100 times, we’re using machine learning techniques to try and assign IDs. For now, we’re using scikit-learn tools to try and match unknown strings to IDs. Specifically, we’re using scikit-learn’s feature_extraction tools for analyzing the known affiliation strings, and the SGDClassifier to generate models. We then pass the unknown strings through the model and extract both a best match and a confidence estimate. The process can be very memory intensive because of the large number of classes – nearly 6600, one for each aff_id. A typical data set of 25,000 unmatched strings takes about an hour on a modest workstation (Apple iMac with an Intel i5 processor and 24 GB of memory).
We’ve found the process is reliable to the limits of the input dictionary, and also found that it’s much more of a data curation problem than a machine learning problem. Curation of a machine learning model isn’t trivial. It’s very sensitive to both errors in assigned IDs (which are rare but do occasionally occur) and to ambiguous affiliation strings or strings that can’t be split cleanly. Examples of the latter include affiliation strings that include (say) both a national laboratory affiliation and a university one (e.g. the Italian INFN centers, NIKHEF member institutions, and US DOE Laboratories), or cases where we have an incomplete mapping of all parent-child relations (e.g. where we have an ID for a university’s Department of Physics, but not a Department of Statistics or Materials Science).
For now, we’re using machine learning to assist with curation, but we’re not yet confident enough to pass it an affiliation string and guarantee it returns the correct ID; it’s not (yet) a part of our hands-off pipeline. It’s an ongoing process of improvement, and always will be as new metadata keeps coming in.
What’s next?
Searching ADS by affiliation is already very useful for helping with disambiguation, and for helping build institutional publication lists. We hope to make it even more powerful by integrating our system with ROR and integrating publisher-specific identifiers in our workflow. We are actively working with other projects to extend ROR identifiers (which are assigned one per institution) to the department level. In addition, we hope to improve the user experience by coupling affiliation with authors, adding hover-over expansion of abbreviations, and implementing auto-complete with the institution search. As always, we welcome feedback and corrections.
 Science Explorer
Science Explorer